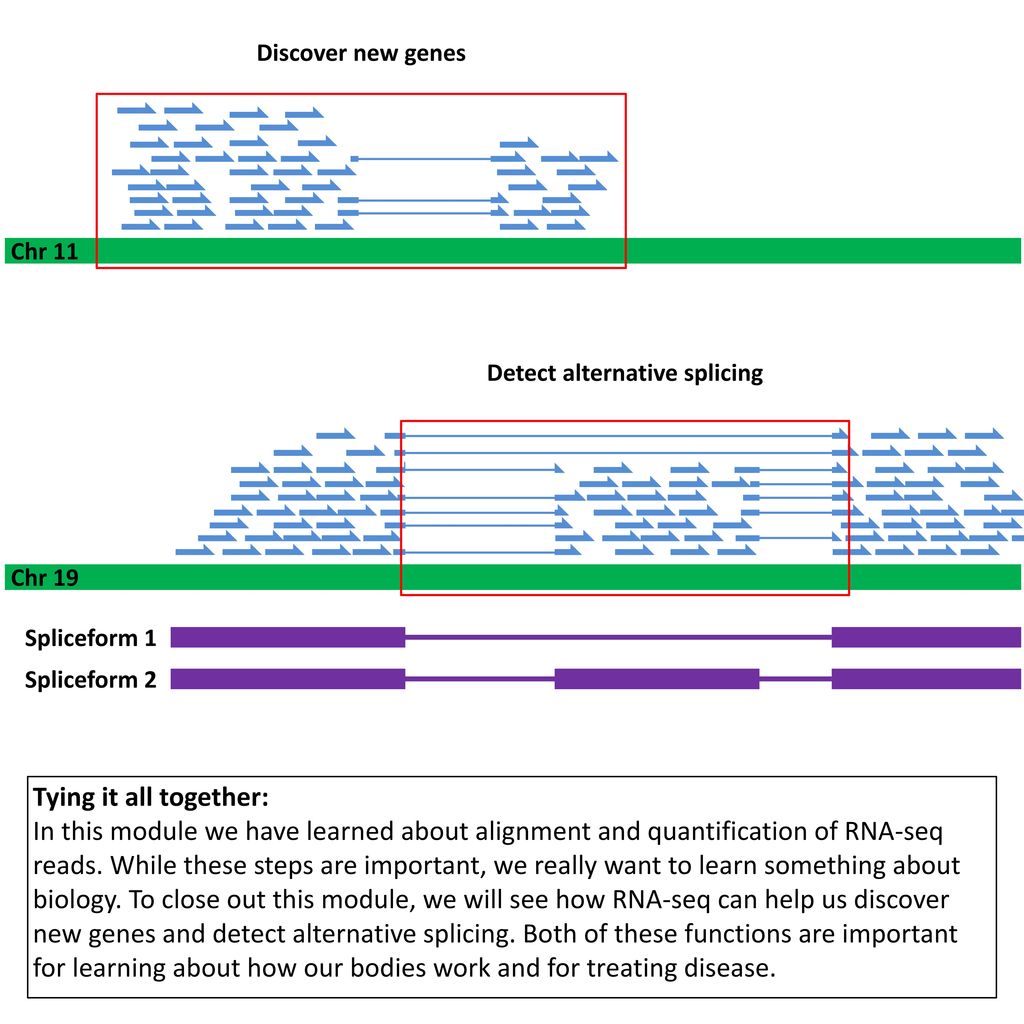
![PDF] Detection, annotation and visualization of alternative splicing from RNA-Seq data with SplicingViewer. | Semantic Scholar PDF] Detection, annotation and visualization of alternative splicing from RNA-Seq data with SplicingViewer. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/d09ff8a3b1debf3f4a38d5ae620cdbc855b9962f/2-Figure1-1.png)
PDF] Detection, annotation and visualization of alternative splicing from RNA-Seq data with SplicingViewer. | Semantic Scholar

EBChangePoint – An empirical Bayes change-point model for identifying 3′ and 5′ alternative splicing by RNA-Seq | RNA-Seq Blog

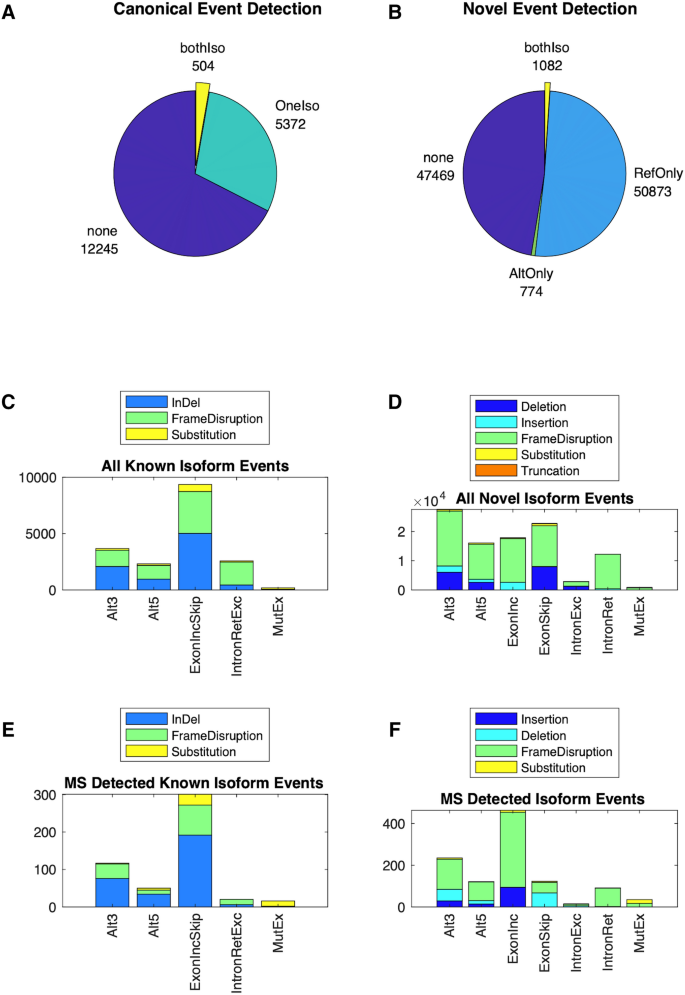
Comparison of RNA-seq and microarray platforms for splice event detection using a cross-platform algorithm | BMC Genomics | Full Text

JCI Insight - The ribosomal prolyl-hydroxylase OGFOD1 decreases during cardiac differentiation and modulates translation and splicing

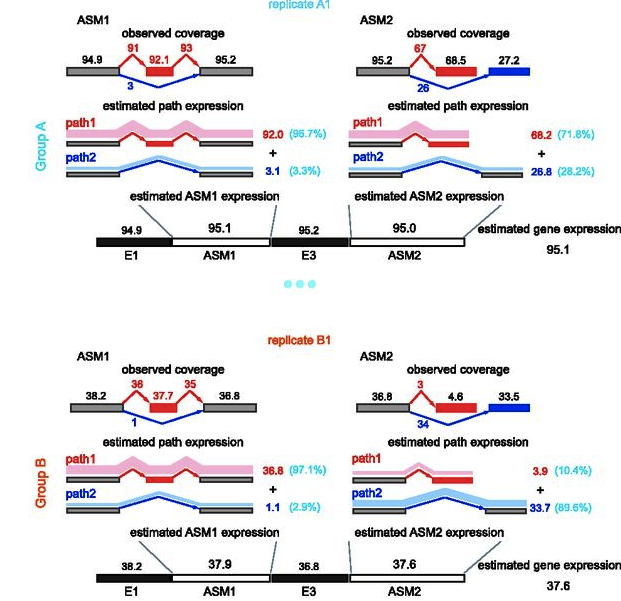
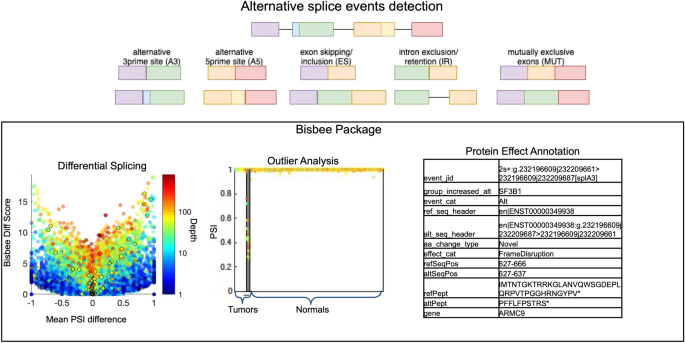
SplAdder – Identification, quantification and testing of alternative splicing events from RNA-Seq data | RNA-Seq Blog

SplAdder – Identification, quantification and testing of alternative splicing events from RNA-Seq data | RNA-Seq Blog

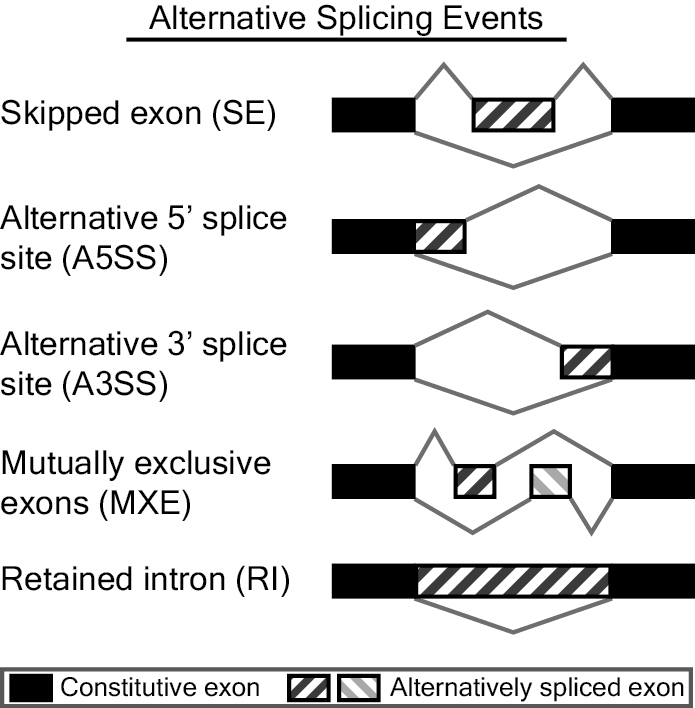
Examples of PS-regulated splicing events. Alternative splicing events... | Download Scientific Diagram

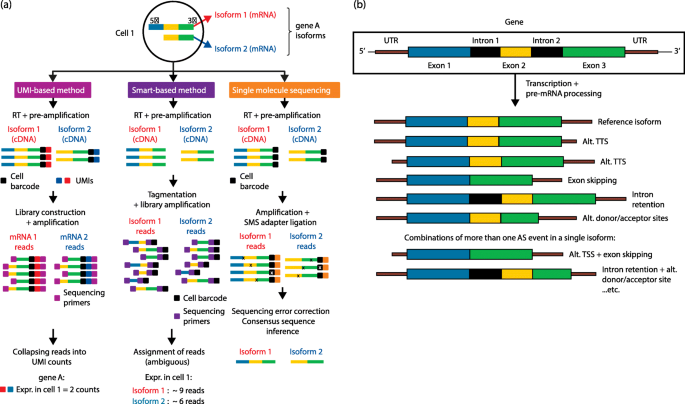
Single-cell differential splicing analysis reveals high heterogeneity of liver tumor-infiltrating T cells | Scientific Reports

Use of RNA-Seq data to detect alternative splicing and exon skipping... | Download Scientific Diagram
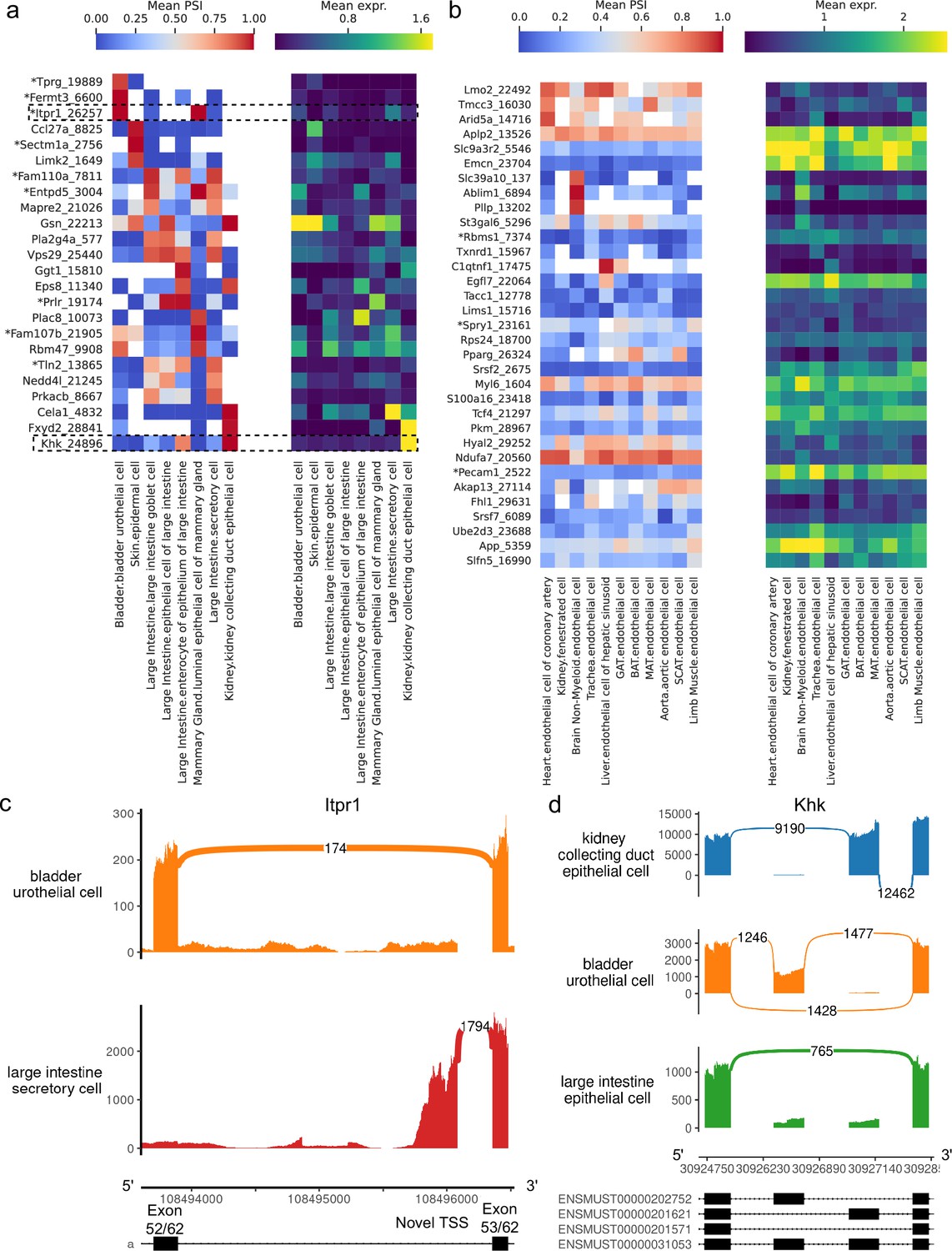
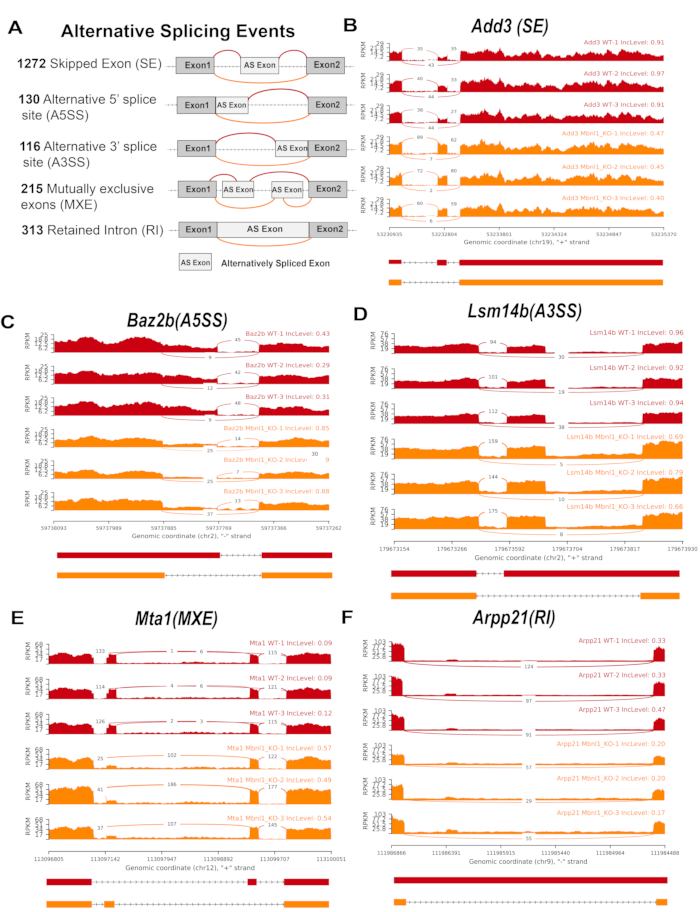
Robust and annotation-free analysis of alternative splicing across diverse cell types in mice | eLife